wbia.algo.graph package¶
Subpackages¶
Submodules¶
wbia.algo.graph.core module¶
-
class
wbia.algo.graph.core.AltConstructors[source]¶ Bases:
object
-
class
wbia.algo.graph.core.AnnotInference(ibs, aids=[], nids=None, autoinit=True, verbose=False)[source]¶ Bases:
utool.util_dev.NiceRepr,wbia.algo.graph.core.AltConstructors,wbia.algo.graph.core.MiscHelpers,wbia.algo.graph.core.Feedback,wbia.algo.graph.core.NameRelabel,wbia.algo.graph.mixin_dynamic.NonDynamicUpdate,wbia.algo.graph.mixin_dynamic.Recovery,wbia.algo.graph.mixin_dynamic.Consistency,wbia.algo.graph.mixin_dynamic.Redundancy,wbia.algo.graph.mixin_dynamic.DynamicUpdate,wbia.algo.graph.mixin_priority.Priority,wbia.algo.graph.mixin_matching.CandidateSearch,wbia.algo.graph.mixin_matching.InfrLearning,wbia.algo.graph.mixin_matching.AnnotInfrMatching,wbia.algo.graph.mixin_helpers.AssertInvariants,wbia.algo.graph.mixin_helpers.DummyEdges,wbia.algo.graph.mixin_helpers.Convenience,wbia.algo.graph.mixin_helpers.AttrAccess,wbia.algo.graph.mixin_simulation.SimulationHelpers,wbia.algo.graph.mixin_loops.InfrReviewers,wbia.algo.graph.mixin_loops.InfrLoops,wbia.algo.graph.mixin_viz.GraphVisualization,wbia.algo.graph.mixin_groundtruth.Groundtruth,wbia.algo.graph.mixin_wbia.IBEISIO,wbia.algo.graph.mixin_wbia.IBEISGroundtruthclass for maintaining state of an identification
Terminology and Concepts:
- CommandLine:
wbia make_qt_graph_interface –show –aids=1,2,3,4,5,6,7 wbia AnnotInference:0 –show wbia AnnotInference:1 –show wbia AnnotInference:2 –show
wbia AnnotInference:0 –loginfr
- Doctest:
>>> from wbia.algo.graph.core import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='PZ_MTEST') >>> aids = [1, 2, 3, 4, 5, 6] >>> infr = AnnotInference(ibs, aids, autoinit=True, verbose=1000) >>> result = ('infr = %s' % (infr,)) >>> print(result) >>> ut.quit_if_noshow() >>> use_image = True >>> infr.initialize_visual_node_attrs() >>> # Note that there are initially no edges >>> infr.show_graph(use_image=use_image) >>> ut.show_if_requested() infr = <AnnotInference(nNodes=6, nEdges=0, nCCs=6)>
Example
>>> # SCRIPT >>> from wbia.algo.graph.core import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='PZ_MTEST') >>> aids = [1, 2, 3, 4, 5, 6, 7, 9] >>> infr = AnnotInference(ibs, aids, autoinit=True) >>> result = ('infr = %s' % (infr,)) >>> print(result) >>> ut.quit_if_noshow() >>> use_image = False >>> infr.initialize_visual_node_attrs() >>> # Note that there are initially no edges >>> infr.show_graph(use_image=use_image) >>> # But we can add nodes between the same names >>> infr.ensure_mst() >>> infr.show_graph(use_image=use_image) >>> # Add some feedback >>> infr.add_feedback((1, 4), NEGTV) >>> infr.apply_feedback_edges() >>> infr.show_graph(use_image=use_image) >>> ut.show_if_requested()
Example
>>> # SCRIPT >>> from wbia.algo.graph.core import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='PZ_MTEST') >>> aids = [1, 2, 3, 4, 5, 6, 7, 9] >>> infr = AnnotInference(ibs, aids, autoinit=True) >>> result = ('infr = %s' % (infr,)) >>> print(result) >>> ut.quit_if_noshow() >>> use_image = False >>> infr.initialize_visual_node_attrs() >>> infr.ensure_mst() >>> # Add some feedback >>> infr.add_feedback((1, 4), NEGTV) >>> try: >>> infr.add_feedback((1, 10), NEGTV) >>> except ValueError: >>> pass >>> try: >>> infr.add_feedback((11, 12), NEGTV) >>> except ValueError: >>> pass >>> infr.apply_feedback_edges() >>> infr.show_graph(use_image=use_image) >>> ut.show_if_requested()
- Ignore:
>>> import wbia >>> import utool as ut >>> ibs = wbia.opendb(defaultdb='PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, 'all') >>> class_ = infr >>> fpath = None >>> static_attrs = ut.check_static_member_vars(class_, fpath) >>> uninitialized = set(infr.__dict__.keys()) - set(static_attrs)
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
subgraph(aids)[source]¶ Makes a new inference object that is a subset of the original.
Note, this is not robust, be careful. The subgraph should be treated as read only. Do not commit any reviews made from here.
-
subparams(prefix)[source]¶ Returns dict of params prefixed with <prefix>. The returned dict does not contain the prefix
- Doctest:
>>> from wbia.algo.graph.core import * >>> import wbia >>> infr = wbia.AnnotInference(None) >>> result = ut.repr2(infr.subparams('refresh')) >>> print(result) {'method': 'binomial', 'patience': 72, 'thresh': 0.052, 'window': 20}
-
class
wbia.algo.graph.core.Feedback[source]¶ Bases:
object-
add_feedback(edge, evidence_decision=None, tags=None, user_id=None, meta_decision=None, confidence=None, timestamp_c1=None, timestamp_c2=None, timestamp_s1=None, timestamp=None, verbose=None, priority=None)[source]¶ - Doctest:
>>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('testdb1') >>> infr.add_feedback((5, 6), POSTV) >>> infr.add_feedback((5, 6), NEGTV, tags=['photobomb']) >>> infr.add_feedback((1, 2), INCMP) >>> print(ut.repr2(infr.internal_feedback, nl=2)) >>> assert len(infr.external_feedback) == 0 >>> assert len(infr.internal_feedback) == 2 >>> assert len(infr.internal_feedback[(5, 6)]) == 2 >>> assert len(infr.internal_feedback[(1, 2)]) == 1
-
apply_feedback_edges()[source]¶ Transforms the feedback dictionaries into nx graph edge attributes
- CommandLine:
- python -m wbia.algo.graph.core apply_feedback_edges
- Doctest:
>>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('testdb1') >>> infr.reset_feedback() >>> infr.params['inference.enabled'] = False >>> #infr.add_feedback((1, 2), 'unknown', tags=[]) >>> infr.add_feedback((1, 2), INCMP, tags=[]) >>> infr.apply_feedback_edges() >>> print('edges = ' + ut.repr4(dict(infr.graph.edges))) >>> result = str(infr) >>> print(result) <AnnotInference(nNodes=6, nEdges=3, nCCs=4)>
-
edge_decision(edge)[source]¶ Gets a decision on an edge, either explicitly or implicitly
- CommandLine:
- python -m wbia.algo.graph.core edge_decision
- Doctest:
>>> from wbia.algo.graph.core import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=1, p_incon=1) >>> decision = infr.edge_decision((1, 2)) >>> print('decision = %r' % (decision,)) >>> assert decision == POSTV >>> decision = infr.edge_decision((199, 299)) >>> print('decision = %r' % (decision,)) >>> assert decision == UNREV
-
feedback_data_keys= ['evidence_decision', 'tags', 'user_id', 'meta_decision', 'timestamp_c1', 'timestamp_c2', 'timestamp_s1', 'timestamp', 'confidence']¶
-
feedback_keys= ['evidence_decision', 'tags', 'user_id', 'meta_decision', 'timestamp_c1', 'timestamp_c2', 'timestamp_s1', 'timestamp', 'confidence', 'num_reviews', 'review_id']¶
-
reset(state='empty')[source]¶ Removes all edges from graph and resets name labels.
- Ignore:
>>> from wbia.algo.graph.core import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=5) >>> assert len(list(infr.edges())) > 0 >>> infr.reset(state='empty') >>> assert len(list(infr.edges())) == 0
-
-
class
wbia.algo.graph.core.MiscHelpers[source]¶ Bases:
object-
add_aids(aids, nids=None)[source]¶ - CommandLine:
- python -m wbia.algo.graph.core add_aids –show
- Doctest:
>>> from wbia.algo.graph.core import * # NOQA >>> aids_ = [1, 2, 3, 4, 5, 6, 7, 9] >>> infr = AnnotInference(ibs=None, aids=aids_, autoinit=True) >>> aids = [2, 22, 7, 9, 8] >>> nids = None >>> infr.add_aids(aids, nids) >>> result = infr.aids >>> print(result) >>> assert len(infr.graph) == len(infr.aids) ... [1, 2, 3, 4, 5, 6, 7, 9, 22, 8]
-
print(msg, level=1, color=None)¶
-
remove_aids(aids)[source]¶ Remove annotations from the graph. :returns: split: indicates which PCCs were split by this action. :rtype: dict
Note
This may cause unintended splits!
- Ignore:
>>> from graphid import demo, util >>> infr = demo.demodata_infr(num_pccs=5, pos_redun=1) >>> infr.refresh_candidate_edges() >>> infr.pin_node_layout() >>> before = infr.copy() >>> aids = infr.aids[::5] >>> splits = infr.remove_aids(aids) >>> assert len(splits['old']) > 0 >>> infr.assert_invariants() >>> # xdoc: +REQUIRES(--show) >>> util.qtensure() >>> after = infr >>> before.show(fnum=1, pnum=(1, 2, 1), pickable=True) >>> after.show(fnum=1, pnum=(1, 2, 2), pickable=True)
-
-
class
wbia.algo.graph.core.NameRelabel[source]¶ Bases:
object-
connected_component_status()[source]¶ Returns: num_inconsistent, num_names_max Return type: dict - CommandLine:
- python -m wbia.algo.graph.core connected_component_status
Example
>>> # DISABLE_DOCTEST >>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('testdb1') >>> infr.add_feedback_from([(2, 3, NEGTV), (5, 6, NEGTV), (1, 2, POSTV)]) >>> status = infr.connected_component_status() >>> print(ut.repr3(status))
-
relabel_using_reviews(graph=None, rectify=True)[source]¶ Relabels nodes in graph based on positive connected components
This will change all of the names on the nodes to be consistent while preserving any existing names as best as possible. If rectify=False, this will be faster, but the old names will not be preserved and each PCC will be assigned an arbitrary name.
Note
if something messes up you can call infr.reset_labels_to_wbia() to reset node labels to their original values — this will almost always put the graph in an inconsistent state — but then you can this with rectify=True to fix everything up.
Parameters: - graph (nx.Graph, optional) – only edges in graph are relabeled defaults to current graph.
- rectify (bool, optional) – if True names attempt to remain consistent otherwise there are no restrictions on name labels other than that they are distinct.
-
wbia.algo.graph.demo module¶
TODO: separate out the tests and make this file just generate the demo data
-
class
wbia.algo.graph.demo.DummyVerif(infr)[source]¶ Bases:
objectgenerates dummy scores between edges (not necesarilly in the graph)
- CommandLine:
- python -m wbia.algo.graph.demo DummyVerif:1
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.demo import * # NOQA >>> from wbia.algo.graph import demo >>> import networkx as nx >>> kwargs = dict(num_pccs=6, p_incon=.5, size_std=2) >>> infr = demo.demodata_infr(**kwargs) >>> infr.dummy_verif.predict_edges([(1, 2)]) >>> infr.dummy_verif.predict_edges([(1, 21)]) >>> assert len(infr.dummy_verif.infr.task_probs['match_state']) == 2
-
dummy_ranker(u, K=10)[source]¶ simulates the ranking algorithm. Order is defined using the dummy vsone scores, but tests are only applied to randomly selected gt and gf pairs. So, you usually will get a gt result, but you might not if all the scores are bad.
-
find_candidate_edges(K=10)[source]¶ Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.demo import * # NOQA >>> from wbia.algo.graph import demo >>> import networkx as nx >>> kwargs = dict(num_pccs=40, size=2) >>> infr = demo.demodata_infr(**kwargs) >>> edges = list(infr.dummy_verif.find_candidate_edges(K=100)) >>> scores = np.array(infr.dummy_verif.predict_edges(edges))
-
predict_proba_df(edges)[source]¶ - CommandLine:
- python -m wbia.algo.graph.demo DummyVerif.predict_edges
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.demo import * # NOQA >>> from wbia.algo.graph import demo >>> import networkx as nx >>> kwargs = dict(num_pccs=40, size=2) >>> infr = demo.demodata_infr(**kwargs) >>> verif = infr.dummy_verif >>> edges = list(infr.graph.edges()) >>> probs = verif.predict_proba_df(edges) >>> #print('scores = %r' % (scores,)) >>> #hashid = ut.hash_data(scores) >>> #print('hashid = %r' % (hashid,)) >>> #assert hashid == 'cdlkytilfeqgmtsihvhqwffmhczqmpil'
-
show_score_probs()[source]¶ - CommandLine:
- python -m wbia.algo.graph.demo DummyVerif.show_score_probs –show
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.demo import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference(None) >>> verif = DummyVerif(infr) >>> verif.show_score_probs() >>> ut.show_if_requested()
-
wbia.algo.graph.demo.demo2()[source]¶ - CommandLine:
- python -m wbia.algo.graph.demo demo2 –viz python -m wbia.algo.graph.demo demo2
Example
>>> # DISABLE_DOCTEST >>> from wbia.algo.graph.demo import * # NOQA >>> result = demo2() >>> print(result)
-
wbia.algo.graph.demo.demodata_infr(**kwargs)[source]¶ kwargs = {}
- CommandLine:
- python -m wbia.algo.graph.demo demodata_infr –show python -m wbia.algo.graph.demo demodata_infr –num_pccs=25 python -m wbia.algo.graph.demo demodata_infr –profile –num_pccs=100
- Ignore:
>>> from wbia.algo.graph.demo import * # NOQA >>> from wbia.algo.graph import demo >>> import networkx as nx >>> kwargs = dict(num_pccs=6, p_incon=.5, size_std=2) >>> kwargs = ut.argparse_dict(kwargs) >>> infr = demo.demodata_infr(**kwargs) >>> pccs = list(infr.positive_components()) >>> assert len(pccs) == kwargs['num_pccs'] >>> nonfull_pccs = [cc for cc in pccs if len(cc) > 1 and nx.is_empty(nx.complement(infr.pos_graph.subgraph(cc)))] >>> expected_n_incon = len(nonfull_pccs) * kwargs['p_incon'] >>> n_incon = len(list(infr.inconsistent_components())) >>> # TODO can test that we our sample num incon agrees with pop mean >>> #sample_mean = n_incon / len(nonfull_pccs) >>> #pop_mean = kwargs['p_incon'] >>> print('status = ' + ut.repr4(infr.status(extended=True))) >>> ut.quit_if_noshow() >>> infr.show(pickable=True, groupby='name_label') >>> ut.show_if_requested()
- Ignore:
- kwargs = {
- ‘ccs’: [[1, 2, 3], [4, 5]]
}
wbia.algo.graph.mixin_dynamic module¶
Todo
Negative bookkeeping, needs a small re-organization fix. MOVE FROM neg_redun_metagraph TO neg_metagraph
Instead of maintaining a graph that contains PCCS which are neg redundant to each other, the graph should maintain PCCs that have ANY negative edge between them (aka 1 neg redundant). Then that edge should store a flag indicating the strength / redundancy of that connection. A better idea might be to store both neg_redun_metagraph AND neg_metagraph.
TODO: this (all neg-redun functionality can be easilly consolidated into the neg-metagraph-update. note, we have to allow inconsistent pccs to be in the neg redun graph, we just filter them out afterwords)
-
class
wbia.algo.graph.mixin_dynamic.Consistency[source]¶ Bases:
object-
consistent_components(graph=None)[source]¶ Generates consistent PCCs. These PCCs contain no internal negative edges.
Yields: cc – set: nodes within the PCC
-
inconsistent_components(graph=None)[source]¶ Generates inconsistent PCCs. These PCCs contain internal negative edges indicating an error exists.
-
is_consistent(cc)[source]¶ Determines if a PCC contains inconsistencies
Parameters: cc (set) – nodes in a PCC Returns: bool: returns True unless cc contains any negative edges Return type: flag Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=1, p_incon=1) >>> assert not infr.is_consistent(next(infr.positive_components())) >>> infr = demo.demodata_infr(num_pccs=1, p_incon=0) >>> assert infr.is_consistent(next(infr.positive_components()))
-
-
class
wbia.algo.graph.mixin_dynamic.DynamicUpdate[source]¶ Bases:
object# 12 total possible states
# details of these states. POSITIVE, WITHIN, CONSISTENT
- pos-within never changes PCC status
- never introduces inconsistency
- might add pos-redun
- POSITIVE, WITHIN, INCONSISTENT
- pos-within never changes PCC status
- might fix inconsistent edge
- POSITIVE, BETWEEN, BOTH_CONSISTENT
- pos-between edge always does merge
- POSITIVE, BETWEEN, ANY_INCONSISTENT
- pos-between edge always does merge
- pos-between never fixes inconsistency
- NEGATIVE, WITHIN, CONSISTENT
- might split PCC, results will be consistent
- might causes an inconsistency
- NEGATIVE, WITHIN, INCONSISTENT
- might split PCC, results may be inconsistent
- NEGATIVE, BETWEEN, BOTH_CONSISTENT
- might add neg-redun
- NEGATIVE, BETWEEN, ANY_INCONSISTENT
- might add to incon-neg-external
- neg-redun not tracked for incon.
- UNINFERABLE, WITHIN, CONSISTENT
- might remove pos-redun
- might split PCC, results will be consistent
- UNINFERABLE, WITHIN, INCONSISTENT
- might split PCC, results may be inconsistent
- UNINFERABLE, BETWEEN, BOTH_CONSISTENT
- might remove neg-redun
- UNINFERABLE, BETWEEN, ANY_INCONSISTENT
- might remove incon-neg-external
-
add_review_edge(edge, decision)[source]¶ Adds edge to the dynamically connected graphs and updates dynamically inferrable edge attributes.
-
ensure_edges_from(edges)[source]¶ Finds edges that don’t exist and adds them as unreviwed edges. Returns new edges that were added.
-
class
wbia.algo.graph.mixin_dynamic.NonDynamicUpdate[source]¶ Bases:
object-
apply_nondynamic_update(graph=None)[source]¶ Recomputes all dynamic bookkeeping for a graph in any state. This ensures that subsequent dyanmic inference can be applied.
- CommandLine:
- python -m wbia.algo.graph.mixin_dynamic apply_nondynamic_update
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> from wbia.algo.graph import demo >>> num_pccs = 250 >>> kwargs = dict(num_pccs=100, p_incon=.3) >>> infr = demo.demodata_infr(infer=False, **kwargs) >>> graph = None >>> infr.apply_nondynamic_update() >>> infr.assert_neg_metagraph()
-
categorize_edges(graph=None, ne_to_edges=None)[source]¶ Non-dynamically computes the status of each edge in the graph. This is can be used to verify the dynamic computations and update when the dynamic state is lost.
- CommandLine:
- python -m wbia.algo.graph.mixin_dynamic categorize_edges –profile
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> from wbia.algo.graph import demo >>> num_pccs = 250 if ut.get_argflag('--profile') else 100 >>> kwargs = dict(num_pccs=100, p_incon=.3) >>> infr = demo.demodata_infr(infer=False, **kwargs) >>> graph = None >>> cat = infr.categorize_edges()
-
collapsed_meta_edges(graph=None)[source]¶ Collapse the grah such that each PCC is a node. Get a list of edges within/between each PCC.
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
-
class
wbia.algo.graph.mixin_dynamic.Recovery[source]¶ Bases:
objectrecovery funcs
-
is_recovering(edge=None)[source]¶ Checks to see if the graph is inconsinsistent.
Parameters: edge (None) – If None, then returns True if the graph contains any inconsistency. Otherwise, returns True if the edge is related to an inconsistent component via a positive or negative connection. Returns: flag Return type: bool - CommandLine:
- python -m wbia.algo.graph.mixin_dynamic is_recovering
- Doctest:
>>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=4, size=4, ignore_pair=True) >>> infr.ensure_cliques(meta_decision=SAME) >>> a, b, c, d = map(list, infr.positive_components()) >>> assert infr.is_recovering() is False >>> infr.add_feedback((a[0], a[1]), NEGTV) >>> assert infr.is_recovering() is True >>> assert infr.is_recovering((a[2], a[3])) is True >>> assert infr.is_recovering((a[3], b[0])) is True >>> assert infr.is_recovering((b[0], b[1])) is False >>> infr.add_feedback((a[3], b[2]), NEGTV) >>> assert infr.is_recovering((b[0], b[1])) is True >>> assert infr.is_recovering((c[0], d[0])) is False >>> infr.add_feedback((b[2], c[0]), NEGTV) >>> assert infr.is_recovering((c[0], d[0])) is False >>> result = ut.repr4({ >>> 'pccs': sorted(list(infr.positive_components())), >>> 'iccs': sorted(list(infr.inconsistent_components())), >>> }, nobr=True, si=True, itemsep='') >>> print(result) iccs: [{1,2,3,4}], pccs: [{5,6,7,8},{9,10,11,12},{13,14,15,16},{1,2,3,4}],
-
-
class
wbia.algo.graph.mixin_dynamic.Redundancy[source]¶ Bases:
wbia.algo.graph.mixin_dynamic._RedundancyComputersmethods for dynamic redundancy book-keeping
-
filter_edges_flagged_as_redun(edges)[source]¶ Returns only edges that are not flagged as redundant. Uses bookkeeping structures
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=1, size=4) >>> infr.clear_edges() >>> infr.ensure_cliques() >>> infr.clear_feedback() >>> print(ut.repr4(infr.status())) >>> nonredun_edges = list(infr.filter_edges_flagged_as_redun( >>> infr.unreviewed_graph.edges())) >>> assert len(nonredun_edges) == 6
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
update_extern_neg_redun(nid, may_add=True, may_remove=True, force=False)[source]¶ Checks if nid is negative redundant to any other cc it has at least one negative review to. (TODO: NEG REDUN CAN BE CONSOLIDATED VIA NEG-META-GRAPH)
-
wbia.algo.graph.mixin_groundtruth module¶
wbia.algo.graph.mixin_helpers module¶
-
class
wbia.algo.graph.mixin_helpers.AttrAccess[source]¶ Bases:
objectContains non-core helper functions
-
gen_edge_attrs(key, edges=None, default=NoParam, on_missing=None)[source]¶ maybe change to gen edge items
-
-
class
wbia.algo.graph.mixin_helpers.Convenience[source]¶ Bases:
object-
incomp_graph¶
-
neg_graph¶
-
pair_connection_info(aid1, aid2)[source]¶ Helps debugging when ibs.nids has info that annotmatch/staging do not
Example
>>> # # FIXME failing-test (22-Jul-2020) GZ_Master1 doesn't exist >>> # xdoctest: +SKIP >>> from wbia.algo.graph.mixin_helpers import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='GZ_Master1') >>> infr = wbia.AnnotInference(ibs, 'all', autoinit=True) >>> infr.reset_feedback('staging', apply=True) >>> infr.relabel_using_reviews(rectify=False) >>> aid1, aid2 = 1349, 3087 >>> aid1, aid2 = 1535, 2549 >>> infr.pair_connection_info(aid1, aid2)
>>> aid1, aid2 = 4055, 4286 >>> aid1, aid2 = 6555, 6882 >>> aid1, aid2 = 712, 803 >>> aid1, aid2 = 3883, 4220 >>> infr.pair_connection_info(aid1, aid2)
-
pos_graph¶
-
unknown_graph¶
-
unreviewed_graph¶
-
-
class
wbia.algo.graph.mixin_helpers.DummyEdges[source]¶ Bases:
object-
ensure_cliques(label='name_label', meta_decision=None)[source]¶ Force each name label to be a clique.
Parameters: - label (str) – node attribute to use as the group id to form the cliques.
- meta_decision (str) – if specified adds clique edges as feedback items with this decision. Otherwise the edges are only explicitly added to the graph.
- infr –
- label – (default = ‘name_label’)
- decision (str) – (default = ‘unreviewed’)
- CommandLine:
- python -m wbia.algo.graph.mixin_helpers ensure_cliques
- Doctest:
>>> from wbia.algo.graph.mixin_helpers import * # NOQA >>> from wbia.algo.graph import demo >>> label = 'name_label' >>> infr = demo.demodata_infr(num_pccs=3, size=5) >>> print(infr.status()) >>> assert infr.status()['nEdges'] < 33 >>> infr.ensure_cliques() >>> print(infr.status()) >>> assert infr.status()['nEdges'] == 33 >>> assert infr.status()['nUnrevEdges'] == 12 >>> assert len(list(infr.find_clique_edges(label))) > 0 >>> infr.ensure_cliques(meta_decision=SAME) >>> assert infr.status()['nUnrevEdges'] == 0 >>> assert len(list(infr.find_clique_edges(label))) == 0
-
ensure_mst(label='name_label', meta_decision='same')[source]¶ Ensures that all names are names are connected.
Parameters: - label (str) – node attribute to use as the group id to form the mst.
- meta_decision (str) – if specified adds clique edges as feedback items with this decision. Otherwise the edges are only explicitly added to the graph. This makes feedback items with user_id=algo:mst and with a confidence of guessing.
- Ignore:
annots = ibs.annots(infr.aids) def fix_name(n):
import re n = re.sub(’ ‘, ‘ ‘, n) return re.sub(’ *-? *BBQ[0-9]’, ‘’, n)ut.fix_embed_globals() new_names = [fix_name(n) for n in annots.names] set(new_names)
annots.names = new_names
infr.set_node_attrs(‘name_fix’, ut.dzip(infr.aids, new_names)) label = ‘name_fix’ infr.ensure_mst(label)
infr.set_node_attrs(‘name_label’, ut.dzip(infr.aids, annots.nids))
- Ignore:
- label = ‘name_label’
- Doctest:
>>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=3, size=4) >>> assert infr.status()['nCCs'] == 3 >>> infr.clear_edges() >>> assert infr.status()['nCCs'] == 12 >>> infr.ensure_mst() >>> assert infr.status()['nCCs'] == 3
- Doctest:
>>> from wbia.algo.graph.mixin_dynamic import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', 'all', autoinit=True) >>> infr.reset_feedback('annotmatch', apply=True) >>> assert infr.status()['nInconsistentCCs'] == 0 >>> assert infr.status()['nCCs'] == 41 >>> label = 'name_label' >>> new_edges = infr.find_mst_edges(label=label) >>> assert len(new_edges) == 0 >>> infr.clear_edges() >>> assert infr.status()['nCCs'] == 119 >>> infr.ensure_mst() >>> assert infr.status()['nCCs'] == 41
-
find_clique_edges(label='name_label')[source]¶ Augmenting edges that would complete each the specified cliques. (based on the group inferred from label)
Parameters: label (str) – node attribute to use as the group id to form the cliques.
-
find_connecting_edges()[source]¶ Searches for a small set of edges, which if reviewed as positive would ensure that each PCC is k-connected. Note that in somes cases this is not possible
-
find_mst_edges(label='name_label')[source]¶ Returns edges to augment existing PCCs (by label) in order to ensure they are connected with positive edges.
- CommandLine:
- python -m wbia.algo.graph.mixin_helpers find_mst_edges –profile
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_helpers import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, 'all', autoinit=True) >>> label = 'orig_name_label' >>> label = 'name_label' >>> infr.find_mst_edges() >>> infr.ensure_mst()
- Ignore:
- old_mst_edges = [
- e for e, d in infr.edges(data=True) if d.get(‘user_id’, None) == ‘algo:mst’
] infr.graph.remove_edges_from(old_mst_edges) infr.pos_graph.remove_edges_from(old_mst_edges) infr.neg_graph.remove_edges_from(old_mst_edges) infr.incomp_graph.remove_edges_from(old_mst_edges)
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
wbia.algo.graph.mixin_loops module¶
-
class
wbia.algo.graph.mixin_loops.InfrLoops[source]¶ Bases:
objectAlgorithm control flow loops
-
hardcase_review_gen()[source]¶ Subiterator for hardcase review
Re-review non-confident edges that vsone did not classify correctly
-
incon_recovery_gen()[source]¶ Subiterator for recovery mode of the mainm algorithm
Iterates until the graph is consistent
Note
inconsistency recovery is implicitly handled by the main algorithm, so other phases do not need to call this explicitly. This exists for the case where the only mode we wish to run is inconsistency recovery.
-
main_gen(max_loops=None, use_refresh=True)[source]¶ The main outer loop.
This function is designed as an iterator that will execute the graph algorithm main loop as automatically as possible, but if user input is needed, it will pause and yield the decision it needs help with. Once feedback is given for this item, you can continue the main loop by calling next. StopIteration is raised once the algorithm is complete.
Parameters: Notes
Different phases of the main loop are implemented as subiterators
- CommandLine:
- python -m wbia.algo.graph.mixin_loops main_gen
- Doctest:
>>> # xdoctest: +REQUIRES(--slow) >>> from wbia.algo.graph.mixin_loops import * >>> from wbia.algo.graph.mixin_simulation import UserOracle >>> import wbia >>> infr = wbia.AnnotInference('testdb1', aids='all', >>> autoinit='staging', verbose=4) >>> infr.params['manual.n_peek'] = 10 >>> infr.params['ranking.ntop'] = 1 >>> infr.oracle = UserOracle(.99, rng=0) >>> infr.simulation_mode = False >>> infr.reset() >>> #infr.load_published() >>> gen = infr.main_gen() >>> while True: >>> try: >>> reviews = next(gen) >>> edge, priority, data = reviews[0] >>> feedback = infr.request_oracle_review(edge) >>> infr.add_feedback(edge, **feedback) >>> except StopIteration: >>> break
-
main_loop(max_loops=None, use_refresh=True)[source]¶ DEPRICATED
use list(infr.main_gen) instead or assert not any(infr.main_gen()) maybe this is fine.
-
neg_redun_gen()[source]¶ Subiterator for phase3 of the main algorithm.
Searches for decisions that would commplete negative redundancy
-
pos_redun_gen()[source]¶ Subiterator for phase2 of the main algorithm.
Searches for decisions that would commplete positive redundancy
- Doctest:
>>> from wbia.algo.graph.mixin_loops import * >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids='all', >>> autoinit='staging', verbose=4) >>> #infr.load_published() >>> gen = infr.pos_redun_gen() >>> feedback = next(gen)
-
-
class
wbia.algo.graph.mixin_loops.InfrReviewers[source]¶ Bases:
object-
emit_manual_review(edge, priority=None)[source]¶ Emits a signal containing edges that need review. The callback should present them to a user, get feedback, and then call on_accpet.
-
qt_review_loop()[source]¶ TODO: The loop parts should be a non-mixin class
Qt review loop entry point
- CommandLine:
- python -m wbia.algo.graph.mixin_loops qt_review_loop –show
Example
>>> # SCRIPT >>> import utool as ut >>> import wbia >>> ibs = wbia.opendb('PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, 'all', autoinit=True) >>> infr.ensure_mst() >>> # Add dummy priorities to each edge >>> infr.set_edge_attrs('prob_match', ut.dzip(infr.edges(), [1])) >>> infr.prioritize('prob_match', infr.edges(), reset=True) >>> infr.params['redun.enabled'] = False >>> win = infr.qt_review_loop() >>> import wbia.guitool as gt >>> gt.qtapp_loop(qwin=win, freq=10)
-
wbia.algo.graph.mixin_matching module¶
-
class
wbia.algo.graph.mixin_matching.AnnotInfrMatching[source]¶ Bases:
objectMethods for running matching algorithms
-
apply_match_edges(review_cfg={})[source]¶ Adds results from one-vs-many rankings as edges in the graph
-
apply_match_scores()[source]¶ Applies precomputed matching scores to edges that already exist in the graph. Typically you should run infr.apply_match_edges() before running this.
- CommandLine:
- python -m wbia.algo.graph.core apply_match_scores –show
Example
>>> # xdoctest: +REQUIRES(--slow) >>> # ENABLE_DOCTEST >>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('PZ_MTEST') >>> infr.exec_matching() >>> infr.apply_match_edges() >>> infr.apply_match_scores() >>> infr.get_edge_attrs('score')
-
exec_matching(qaids=None, daids=None, prog_hook=None, cfgdict=None, name_method='node', use_cache=True, invalidate_supercache=False, batch_size=None, ranks_top=5)[source]¶ Loads chip matches into the inference structure Uses graph name labeling and ignores wbia labeling
-
exec_vsone_subset(edges, prog_hook=None)[source]¶ Parameters: prog_hook (None) – (default = None) - CommandLine:
- python -m wbia.algo.graph.core exec_vsone_subset
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('testdb1') >>> infr.ensure_full() >>> edges = [(1, 2), (2, 3)] >>> result = infr.exec_vsone_subset(edges) >>> print(result)
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
-
class
wbia.algo.graph.mixin_matching.CandidateSearch[source]¶ Bases:
wbia.algo.graph.mixin_matching._RedundancyAugmentationSearch for candidate edges
-
ensure_priority_scores(priority_edges)[source]¶ Ensures that priority attributes are assigned to the edges. This does not change the state of the queue.
- Doctest:
>>> import wbia >>> ibs = wbia.opendb('PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, aids='all') >>> infr.ensure_mst() >>> priority_edges = list(infr.edges())[0:1] >>> infr.ensure_priority_scores(priority_edges)
- Doctest:
>>> import wbia >>> ibs = wbia.opendb('PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, aids='all') >>> infr.ensure_mst() >>> # infr.load_published() >>> priority_edges = list(infr.edges()) >>> infr.ensure_priority_scores(priority_edges)
- Doctest:
>>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=6, p_incon=.5, size_std=2) >>> edges = list(infr.edges()) >>> infr.ensure_priority_scores(edges)
-
ensure_task_probs(edges)[source]¶ Ensures that probabilities are assigned to the edges. This gaurentees that infr.task_probs contains data for edges. (Currently only the primary task is actually ensured)
- CommandLine:
- python -m wbia.algo.graph.mixin_matching ensure_task_probs
- Doctest:
>>> # DISABLE_DOCTEST >>> from wbia.algo.graph.mixin_matching import * >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids='all', >>> autoinit='staging') >>> edges = list(infr.edges())[0:3] >>> infr.load_published() >>> assert len(infr.task_probs['match_state']) == 0 >>> infr.ensure_task_probs(edges) >>> assert len(infr.task_probs['match_state']) == 3 >>> infr.ensure_task_probs(edges) >>> assert len(infr.task_probs['match_state']) == 3
- Doctest:
>>> # DISABLE_DOCTEST >>> from wbia.algo.graph.mixin_matching import * >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=6, p_incon=.5, size_std=2) >>> edges = list(infr.edges()) >>> infr.ensure_task_probs(edges) >>> assert all([np.isclose(sum(p.values()), 1) >>> for p in infr.task_probs['match_state'].values()])
-
find_lnbnn_candidate_edges(desired_states=['unreviewed'], can_match_samename=False, can_match_sameimg=False, K=5, Knorm=5, requery=True, prescore_method='csum', score_method='csum', sv_on=True, cfgdict_=None, batch_size=None)[source]¶ Example
>>> # DISABLE_DOCTEST >>> # xdoctest: +REQUIRES(--slow) >>> from wbia.algo.graph import demo >>> infr = demo.demodata_mtest_infr() >>> cand_edges = infr.find_lnbnn_candidate_edges() >>> assert len(cand_edges) > 200, len(cand_edges)
-
-
class
wbia.algo.graph.mixin_matching.InfrLearning[source]¶ Bases:
object-
learn_deploy_verifiers(publish=False)[source]¶ Uses current knowledge to train verifiers for new unseen pairs.
Example
>>> # DISABLE_DOCTEST >>> import wbia >>> ibs = wbia.opendb('PZ_MTEST') >>> infr = wbia.AnnotInference(ibs, aids='all') >>> infr.ensure_mst() >>> publish = False >>> infr.learn_deploy_verifiers()
- Ignore:
- publish = True
-
learn_evaluation_verifiers()[source]¶ Creates a cross-validated ensemble of classifiers to evaluate verifier error cases and groundtruth errors.
- CommandLine:
- python -m wbia.algo.graph.mixin_matching learn_evaluation_verifiers
- Doctest:
>>> # xdoctest: +REQUIRES(module:wbia_cnn, --slow) >>> import wbia >>> infr = wbia.AnnotInference( >>> 'PZ_MTEST', aids='all', autoinit='annotmatch', >>> verbose=4) >>> verifiers = infr.learn_evaluation_verifiers() >>> edges = list(infr.edges()) >>> verif = verifiers['match_state'] >>> probs = verif.predict_proba_df(edges) >>> print(probs)
-
wbia.algo.graph.mixin_priority module¶
-
class
wbia.algo.graph.mixin_priority.Priority[source]¶ Bases:
objectHandles prioritization of edges for review.
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_priority import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=20)
-
confidently_connected(u, v, thresh=2)[source]¶ Checks if u and v are conneted by edges above a confidence threshold
-
confidently_separated(u, v, thresh=2)[source]¶ Checks if u and v are conneted by edges above a confidence threshold
- Doctest:
>>> from wbia.algo.graph.mixin_priority import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.make_demo_infr(ccs=[(1, 2), (3, 4), (5, 6), (7, 8)]) >>> infr.add_feedback((1, 5), NEGTV) >>> infr.add_feedback((5, 8), NEGTV) >>> infr.add_feedback((6, 3), NEGTV) >>> u, v = (1, 4) >>> thresh = 0 >>> assert not infr.confidently_separated(u, v, thresh) >>> infr.add_feedback((2, 3), NEGTV) >>> assert not infr.confidently_separated(u, v, thresh)
-
generate_reviews(pos_redun=None, neg_redun=None, data=False)[source]¶ Dynamic generator that yeilds high priority reviews
-
peek_many(n)[source]¶ Peeks at the top n edges in the queue.
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_priority import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=7, size=5) >>> infr.refresh_candidate_edges() >>> infr.peek_many(50)
-
pop()[source]¶ Main interface to the priority queue used by the algorithm loops. Pops the highest priority edge from the queue.
-
prioritize(metric=None, edges=None, scores=None, force_inconsistent=True, reset=False)[source]¶ Adds edges to the priority queue
- Doctest:
>>> from wbia.algo.graph.mixin_priority import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=7, size=5) >>> infr.ensure_cliques(meta_decision=SAME) >>> # Add a negative edge inside a PCC >>> ccs = list(infr.positive_components()) >>> edge1 = tuple(list(ccs[0])[0:2]) >>> edge2 = tuple(list(ccs[1])[0:2]) >>> infr.add_feedback(edge1, NEGTV) >>> infr.add_feedback(edge2, NEGTV) >>> num_new = infr.prioritize(reset=True) >>> order = infr._peek_many(np.inf) >>> scores = ut.take_column(order, 1) >>> assert scores[0] > 10 >>> assert len(scores) == num_new, 'should prioritize two hypotheis edges' >>> unrev_edges = set(infr.unreviewed_graph.edges()) >>> err_edges = set(ut.flatten(infr.nid_to_errors.values())) >>> edges = set(list(unrev_edges - err_edges)[0:2]) >>> edges.update(list(err_edges)[0:2]) >>> num_new = infr.prioritize(edges=edges, reset=True) >>> order2 = infr._peek_many(np.inf) >>> scores2 = np.array(ut.take_column(order2, 1)) >>> assert np.all(scores2[0:2] > 10) >>> assert np.all(scores2[2:] < 10)
Example
import wbia infr = wbia.AnnotInference(‘PZ_MTEST’, aids=’all’, autoinit=’staging’) infr.verbose = 1000 infr.load_published() incon_edges = set(ut.iflatten(infr.nid_to_errors.values())) assert len(incon_edges) > 0 edges = list(infr.find_pos_redun_candidate_edges()) assert len(set(incon_edges).intersection(set(edges))) == 0 infr.add_candidate_edges(edges)
infr.prioritize() logger.info(ut.repr4(infr.status()))
-
wbia.algo.graph.mixin_simulation module¶
Mixin functionality for experiments, tests, and simulations. This includes recordings measures used to generate plots in JC’s thesis.
wbia.algo.graph.mixin_viz module¶
-
class
wbia.algo.graph.mixin_viz.GraphVisualization[source]¶ Bases:
objectcontains plotting related code
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
show(graph=None, use_image=False, update_attrs=True, with_colorbar=False, pnum=(1, 1, 1), zoomable=True, pickable=False, **kwargs)¶ Parameters: - infr –
- graph (None) – (default = None)
- use_image (bool) – (default = False)
- update_attrs (bool) – (default = True)
- with_colorbar (bool) – (default = False)
- pnum (tuple) – plot number(default = (1, 1, 1))
- zoomable (bool) – (default = True)
- pickable (bool) – (de = False)
- **kwargs – verbose, with_labels, fnum, layout, ax, pos, img_dict, title, layoutkw, framewidth, modify_ax, as_directed, hacknoedge, hacknode, node_labels, arrow_width, fontsize, fontweight, fontname, fontfamilty, fontproperties
- CommandLine:
- python -m wbia.algo.graph.mixin_viz GraphVisualization.show_graph –show
Example
>>> # xdoctest: +REQUIRES(module:pygraphviz) >>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_viz import * # NOQA >>> from wbia.algo.graph import demo >>> import wbia.plottool as pt >>> infr = demo.demodata_infr(ccs=ut.estarmap( >>> range, [(1, 6), (6, 10), (10, 13), (13, 15), (15, 16), >>> (17, 20)])) >>> pnum_ = pt.make_pnum_nextgen(nRows=1, nCols=3) >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> infr.add_feedback((1, 5), INCMP) >>> infr.add_feedback((14, 18), INCMP) >>> infr.refresh_candidate_edges() >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> infr.add_feedback((17, 18), NEGTV) # add inconsistency >>> infr.apply_nondynamic_update() >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> ut.show_if_requested()
-
show_graph(graph=None, use_image=False, update_attrs=True, with_colorbar=False, pnum=(1, 1, 1), zoomable=True, pickable=False, **kwargs)[source]¶ Parameters: - infr –
- graph (None) – (default = None)
- use_image (bool) – (default = False)
- update_attrs (bool) – (default = True)
- with_colorbar (bool) – (default = False)
- pnum (tuple) – plot number(default = (1, 1, 1))
- zoomable (bool) – (default = True)
- pickable (bool) – (de = False)
- **kwargs – verbose, with_labels, fnum, layout, ax, pos, img_dict, title, layoutkw, framewidth, modify_ax, as_directed, hacknoedge, hacknode, node_labels, arrow_width, fontsize, fontweight, fontname, fontfamilty, fontproperties
- CommandLine:
- python -m wbia.algo.graph.mixin_viz GraphVisualization.show_graph –show
Example
>>> # xdoctest: +REQUIRES(module:pygraphviz) >>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_viz import * # NOQA >>> from wbia.algo.graph import demo >>> import wbia.plottool as pt >>> infr = demo.demodata_infr(ccs=ut.estarmap( >>> range, [(1, 6), (6, 10), (10, 13), (13, 15), (15, 16), >>> (17, 20)])) >>> pnum_ = pt.make_pnum_nextgen(nRows=1, nCols=3) >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> infr.add_feedback((1, 5), INCMP) >>> infr.add_feedback((14, 18), INCMP) >>> infr.refresh_candidate_edges() >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> infr.add_feedback((17, 18), NEGTV) # add inconsistency >>> infr.apply_nondynamic_update() >>> infr.show_graph(show_cand=True, simple_labels=True, pickable=True, fnum=1, pnum=pnum_()) >>> ut.show_if_requested()
-
update_visual_attrs(graph=None, show_reviewed_edges=True, show_unreviewed_edges=False, show_inferred_diff=True, show_inferred_same=True, show_recent_review=False, highlight_reviews=True, show_inconsistency=True, wavy=False, simple_labels=False, show_labels=True, reposition=True, use_image=False, edge_overrides=None, node_overrides=None, colorby='name_label', **kwargs)[source]¶
-
visual_edge_attrs¶ all edge visual attrs
-
visual_edge_attrs_appearance¶ attrs that pertain to edge color and style
-
visual_edge_attrs_space¶ attrs that pertain to edge positioning in a plot
-
visual_node_attrs¶
-
wbia.algo.graph.mixin_wbia module¶
-
class
wbia.algo.graph.mixin_wbia.IBEISGroundtruth[source]¶ Bases:
objectMethods for generating training labels for classifiers
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
wbia_guess_if_comparable(aid_pairs)[source]¶ Takes a guess as to which annots are not comparable based on scores and viewpoints. If either viewpoints is null assume they are comparable.
-
-
class
wbia.algo.graph.mixin_wbia.IBEISIO[source]¶ Bases:
objectDirect interface into wbia tables and delta statistics
-
find_unjustified_splits()[source]¶ >>> # ENABLE_DOCTEST >>> from wbia.algo.graph.mixin_helpers import * # NOQA >>> import wbia >>> ibs = wbia.opendb(defaultdb='GZ_Master1') >>> ibs = wbia.opendb(defaultdb='PZ_Master1') >>> infr = wbia.AnnotInference(ibs, 'all', autoinit=True) >>> infr.reset_feedback('staging', apply=True) >>> infr.relabel_using_reviews(rectify=False) >>> unjustified = infr.find_unjustified_splits() >>> review_edges = [] >>> for cc1, cc2 in unjustified: >>> u = next(iter(cc1)) >>> v = next(iter(cc2)) >>> review_edges.append(nxu.e_(u, v)) >>> infr.verbose = 100 >>> infr.prioritize( >>> edges=review_edges, scores=[1] * len(review_edges), >>> reset=True, >>> ) >>> infr.qt_review_loop()
-
get_wbia_name_delta(ignore_unknown=True, relabel=True)[source]¶ Rectifies internal name_labels with the names stored in the name table.
Return a pandas dataframe indicating which names have changed for what annotations.
Parameters: Returns: - pd.DataFrame - name_delta_df - data frame where each row specifies
an aid and its old_name which is in the wbia database and the new_name which is what we infer it should be renamed to.
Example
infr.write_wbia_name_assignment
- CommandLine:
- python -m wbia.algo.graph.mixin_wbia get_wbia_name_delta
- Doctest:
>>> from wbia.algo.graph.mixin_wbia import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids=list(range(1, 10)), >>> autoinit='annotmatch', verbose=4) >>> pccs1 = list(infr.positive_components()) >>> print('pccs1 = %r' % (pccs1,)) >>> print('names = {}'.format(list(infr.gen_node_values('name_label', infr.aids)))) >>> assert pccs1 == [{1, 2, 3, 4}, {5, 6, 7, 8}, {9}] >>> # Split a PCC and then merge two other PCCs >>> infr.add_feedback_from([(1, 2), (1, 3), (1, 4)], evidence_decision=NEGTV) >>> infr.add_feedback((6, 7), NEGTV) >>> infr.add_feedback((5, 8), NEGTV) >>> infr.add_feedback((4, 5), POSTV) >>> infr.add_feedback((7, 8), POSTV) >>> pccs2 = list(infr.positive_components()) >>> print('pccs2 = %r' % (pccs2,)) >>> pccs2 = sorted(pccs2) >>> assert pccs2 == [{9}, {1}, {2, 3, 4, 5, 6}, {7, 8}] >>> print(list(infr.gen_node_values('name_label', infr.aids))) >>> name_delta_df = infr.get_wbia_name_delta() >>> result = str(name_delta_df) >>> print(result) old_name new_name aid 1 06_410 IBEIS_UNKNOWN_0042 5 07_061 06_410 6 07_061 06_410
- Doctest:
>>> from wbia.algo.graph.mixin_wbia import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids=list(range(1, 10)), >>> autoinit='annotmatch', verbose=4) >>> infr.add_feedback_from([(1, 2), (1, 3), (1, 4)], evidence_decision=NEGTV) >>> infr.add_feedback((4, 5), POSTV) >>> name_delta_df = infr.get_wbia_name_delta() >>> result = str(name_delta_df) >>> print(result) old_name new_name aid 2 06_410 07_061 3 06_410 07_061 4 06_410 07_061
- Doctest:
>>> from wbia.algo.graph.mixin_wbia import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids=list(range(1, 10)), >>> autoinit='annotmatch', verbose=4) >>> name_delta_df = infr.get_wbia_name_delta() >>> result = str(name_delta_df) >>> print(result) Empty DataFrame Columns: [old_name, new_name] Index: []
-
match_state_delta(old='annotmatch', new='all')[source]¶ Returns information about state change of annotmatches
By default this will return a pandas dataframe indicating which edges in the annotmatch table have changed and all new edges relative to the current infr.graph state.
Notes
valid values for old and new are {‘annotmatch’, ‘staging’, ‘all’, ‘internal’, or ‘external’}.
The args old/new=’all’ resolves to the internal graph state, ‘annotmatch’ resolves to the on-disk annotmatch table, and ‘staging’ resolves to the on-disk staging table (you can further separate all by specifying ‘internal’ or ‘external’). You any of these old/new combinations to check differences in the state. However, the default values are what you use to sync the graph state to annotmatch.
Parameters: Returns: - pd.DataFrame - edge_delta_df - indicates the old and new values
of the changed edge attributes.
- CommandLine:
- python -m wbia.algo.graph.core match_state_delta
- Doctest:
>>> from wbia.algo.graph.mixin_wbia import * # NOQA >>> import wbia >>> infr = wbia.AnnotInference('PZ_MTEST', aids=list(range(1, 10)), >>> autoinit='annotmatch', verbose=4) >>> # Split a PCC and then merge two other PCCs >>> infr.add_feedback((1, 2), NEGTV) >>> infr.add_feedback((6, 7), NEGTV) >>> infr.add_feedback((5, 8), NEGTV) >>> infr.add_feedback((4, 5), POSTV) >>> infr.add_feedback((7, 8), POSTV) >>> edge_delta_df = infr.match_state_delta() >>> subset = edge_delta_df[['old_evidence_decision', 'new_evidence_decision']] >>> result = str(subset) >>> # sort result by aid1 >>> result = '\n'.join(result.splitlines()[:2] + sorted(result.splitlines()[2:])) >>> print(result) old_evidence_decision new_evidence_decision aid1 aid2 1 2 match nomatch 4 5 NaN match 5 8 unreviewed nomatch 6 7 unreviewed nomatch 7 8 match match
-
name_label_group_delta_info()[source]¶ If the name labeling delta is non-zero then you need to rectify names
infr.relabel_using_reviews(rectify=False)
-
read_wbia_annotmatch_feedback(edges=None)[source]¶ Reads feedback from annotmatch table and returns the result. Internal state is not changed.
Parameters: only_existing_edges (bool) – if True only reads info existing edges - CommandLine:
- python -m wbia.algo.graph.core read_wbia_annotmatch_feedback
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.core import * # NOQA >>> infr = testdata_infr('testdb1') >>> feedback = infr.read_wbia_annotmatch_feedback() >>> items = feedback[(2, 3)] >>> result = ('feedback = %s' % (ut.repr2(feedback, nl=2),)) >>> print(result) >>> assert len(feedback) >= 2, 'should contain at least 2 edges' >>> assert len(items) == 1, '2-3 should have one review' >>> assert items[0]['evidence_decision'] == POSTV, '2-3 must match'
-
read_wbia_staging_feedback(edges=None)[source]¶ Reads feedback from review staging table.
Parameters: infr – Returns: feedback Return type: ? - CommandLine:
- python -m wbia.algo.graph.mixin_wbia read_wbia_staging_feedback
Example
>>> # DISABLE_DOCTEST >>> from wbia.algo.graph.mixin_wbia import * # NOQA >>> import wbia >>> ibs = wbia.opendb('GZ_Master1') >>> infr = wbia.AnnotInference(ibs=ibs, aids='all') >>> feedback = infr.read_wbia_staging_feedback() >>> result = ('feedback = %s' % (ut.repr2(feedback),)) >>> print(result)
-
rrr(verbose=True, reload_module=True)¶ special class reloading function This function is often injected as rrr of classes
-
write_wbia_annotmatch_feedback(edge_delta_df=None)[source]¶ Commits the current state in external and internal into the annotmatch table. Annotmatch only stores the final review in the history of reviews.
By default this will sync the current graph state to the annotmatch table. It computes the edge_delta under the hood, so if you already made one then you can pass it in for a little extra speed.
Parameters: edge_delta_df (pd.DataFrame) – precomputed using match_state_delta. if None it will be computed under the hood.
-
write_wbia_name_assignment(name_delta_df=None, **kwargs)[source]¶ Write the name delta to the annotations table.
It computes the name delta under the hood, so if you already made one then you can pass it in for a little extra speed.
Note
This will call infr.relabel_using_reviews(rectify=True) if name_delta_df is not given directly.
Parameters: name_delta_df (pd.DataFrame) – if None, the value is computed using get_wbia_name_delta. Note you should ensure this delta is made after nodes have been relabeled using reviews.
-
write_wbia_staging_feedback()[source]¶ Commit all reviews in internal_feedback into the staging table. The edges are removed from interal_feedback and added to external feedback. The staging tables stores each review in the order it happened so history is fully reconstructable if staging is never deleted.
This write function is done using the implicit delta maintained by infr.internal_feedback. Therefore, it take no args. This is generally called automatically by infr.accept.
-
wbia.algo.graph.nx_dynamic_graph module¶
-
class
wbia.algo.graph.nx_dynamic_graph.DynConnGraph(*args, **kwargs)[source]¶ Bases:
networkx.classes.graph.Graph,wbia.algo.graph.nx_dynamic_graph.GraphHelperMixinDynamically connected graph.
Maintains a data structure parallel to a normal networkx graph that maintains dynamic connectivity for fast connected compoment queries.
Underlying Data Structures and limitations are
- UnionFind | lg(n) | n | No
- UnionFind2 | n* | n | 1
- EulerTourForest | lg^2(n) | lg^2(n) | lg(n) / lglg(n) - - Ammortized
- it seems to be very quick
References
https://courses.csail.mit.edu/6.851/spring14/lectures/L20.pdf https://courses.csail.mit.edu/6.851/spring14/lectures/L20.html http://cs.stackexchange.com/questions/33595/maintaining-connecte https://en.wikipedia.org/wiki/Dynamic_connectivity#Fully_dynamic_connectivity
- CommandLine:
- python -m wbia.algo.graph.nx_dynamic_graph DynConnGraph
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> self.add_edges_from([(10, 20), (20, 30), (40, 50), (60, 70), (70, 40)]) >>> self._ccs >>> u, v = 20, 1 >>> assert self.node_label(u) != self.node_label(v) >>> assert self.connected_to(u) != self.connected_to(v) >>> self.add_edge(u, v) >>> assert self.node_label(u) == self.node_label(v) >>> assert self.connected_to(u) == self.connected_to(v) >>> self.remove_edge(u, v) >>> assert self.node_label(u) != self.node_label(v) >>> assert self.connected_to(u) != self.connected_to(v) >>> ccs = list(self.connected_components()) >>> ut.quit_if_noshow() >>> import wbia.plottool as pt >>> pt.qtensure() >>> pt.show_nx(self)
# todo: check if nodes exist when adding
-
add_edge(u, v, **attr)[source]¶ Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}} >>> self.add_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3, 4, 5, 6, 7}}
-
add_edges_from(ebunch, **attr)[source]¶ Add all the edges in ebunch_to_add.
Parameters: - ebunch_to_add (container of edges) – Each edge given in the container will be added to the graph. The edges must be given as 2-tuples (u, v) or 3-tuples (u, v, d) where d is a dictionary containing edge data.
- attr (keyword arguments, optional) – Edge data (or labels or objects) can be assigned using keyword arguments.
See also
add_edge()- add a single edge
add_weighted_edges_from()- convenient way to add weighted edges
Notes
Adding the same edge twice has no effect but any edge data will be updated when each duplicate edge is added.
Edge attributes specified in an ebunch take precedence over attributes specified via keyword arguments.
Examples
>>> G = nx.Graph() # or DiGraph, MultiGraph, MultiDiGraph, etc >>> G.add_edges_from([(0, 1), (1, 2)]) # using a list of edge tuples >>> e = zip(range(0, 3), range(1, 4)) >>> G.add_edges_from(e) # Add the path graph 0-1-2-3
Associate data to edges
>>> G.add_edges_from([(1, 2), (2, 3)], weight=3) >>> G.add_edges_from([(3, 4), (1, 4)], label="WN2898")
-
add_node(n, **attr)[source]¶ Add a single node node_for_adding and update node attributes.
Parameters: - node_for_adding (node) – A node can be any hashable Python object except None.
- attr (keyword arguments, optional) – Set or change node attributes using key=value.
See also
Examples
>>> G = nx.Graph() # or DiGraph, MultiGraph, MultiDiGraph, etc >>> G.add_node(1) >>> G.add_node("Hello") >>> K3 = nx.Graph([(0, 1), (1, 2), (2, 0)]) >>> G.add_node(K3) >>> G.number_of_nodes() 3
Use keywords set/change node attributes:
>>> G.add_node(1, size=10) >>> G.add_node(3, weight=0.4, UTM=("13S", 382871, 3972649))
Notes
A hashable object is one that can be used as a key in a Python dictionary. This includes strings, numbers, tuples of strings and numbers, etc.
On many platforms hashable items also include mutables such as NetworkX Graphs, though one should be careful that the hash doesn’t change on mutables.
-
add_nodes_from(nodes, **attr)[source]¶ Add multiple nodes.
Parameters: - nodes_for_adding (iterable container) – A container of nodes (list, dict, set, etc.). OR A container of (node, attribute dict) tuples. Node attributes are updated using the attribute dict.
- attr (keyword arguments, optional (default= no attributes)) – Update attributes for all nodes in nodes. Node attributes specified in nodes as a tuple take precedence over attributes specified via keyword arguments.
See also
Examples
>>> G = nx.Graph() # or DiGraph, MultiGraph, MultiDiGraph, etc >>> G.add_nodes_from("Hello") >>> K3 = nx.Graph([(0, 1), (1, 2), (2, 0)]) >>> G.add_nodes_from(K3) >>> sorted(G.nodes(), key=str) [0, 1, 2, 'H', 'e', 'l', 'o']
Use keywords to update specific node attributes for every node.
>>> G.add_nodes_from([1, 2], size=10) >>> G.add_nodes_from([3, 4], weight=0.4)
Use (node, attrdict) tuples to update attributes for specific nodes.
>>> G.add_nodes_from([(1, dict(size=11)), (2, {"color": "blue"})]) >>> G.nodes[1]["size"] 11 >>> H = nx.Graph() >>> H.add_nodes_from(G.nodes(data=True)) >>> H.nodes[1]["size"] 11
-
clear()[source]¶ Remove all nodes and edges from the graph.
This also removes the name, and all graph, node, and edge attributes.
Examples
>>> G = nx.path_graph(4) # or DiGraph, MultiGraph, MultiDiGraph, etc >>> G.clear() >>> list(G.nodes) [] >>> list(G.edges) []
-
component_nodes(label)¶
-
connected_components()[source]¶ Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7)]) >>> ccs = list(self.connected_components()) >>> result = 'ccs = {}'.format(ut.repr2(ccs, nl=0)) >>> print(result) ccs = [{1, 2, 3}, {4, 5}, {6, 7}]
-
node_label(node)[source]¶ Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7)]) >>> assert self.node_label(2) == self.node_label(1) >>> assert self.node_label(2) != self.node_label(4)
-
remove_edge(u, v)[source]¶ Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}} >>> self.add_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3, 4, 5, 6, 7}} >>> self.remove_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}}
-
remove_edges_from(ebunch)[source]¶ Remove all edges specified in ebunch.
Parameters: ebunch (list or container of edge tuples) – Each edge given in the list or container will be removed from the graph. The edges can be:
- 2-tuples (u, v) edge between u and v.
- 3-tuples (u, v, k) where k is ignored.
See also
remove_edge()- remove a single edge
Notes
Will fail silently if an edge in ebunch is not in the graph.
Examples
>>> G = nx.path_graph(4) # or DiGraph, MultiGraph, MultiDiGraph, etc >>> ebunch = [(1, 2), (2, 3)] >>> G.remove_edges_from(ebunch)
-
remove_node(n)[source]¶ - CommandLine:
- python -m wbia.algo.graph.nx_dynamic_graph remove_node
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_dynamic_graph import * # NOQA >>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (5, 6), (6, 7), (7, 8), (8, 9)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7, 8, 9}} >>> self.remove_node(2) >>> assert self._ccs == {1: {1}, 3: {3}, 4: {4, 5, 6, 7, 8, 9}} >>> self.remove_node(7) >>> assert self._ccs == {1: {1}, 3: {3}, 4: {4, 5, 6}, 8: {8, 9}}
-
remove_nodes_from(nodes)[source]¶ Remove multiple nodes.
Parameters: nodes (iterable container) – A container of nodes (list, dict, set, etc.). If a node in the container is not in the graph it is silently ignored. See also
Examples
>>> G = nx.path_graph(3) # or DiGraph, MultiGraph, MultiDiGraph, etc >>> e = list(G.nodes) >>> e [0, 1, 2] >>> G.remove_nodes_from(e) >>> list(G.nodes) []
-
subgraph(nbunch, dynamic=False)[source]¶ Returns a SubGraph view of the subgraph induced on nodes.
The induced subgraph of the graph contains the nodes in nodes and the edges between those nodes.
Parameters: nodes (list, iterable) – A container of nodes which will be iterated through once. Returns: G – A subgraph view of the graph. The graph structure cannot be changed but node/edge attributes can and are shared with the original graph. Return type: SubGraph View Notes
The graph, edge and node attributes are shared with the original graph. Changes to the graph structure is ruled out by the view, but changes to attributes are reflected in the original graph.
To create a subgraph with its own copy of the edge/node attributes use: G.subgraph(nodes).copy()
For an inplace reduction of a graph to a subgraph you can remove nodes: G.remove_nodes_from([n for n in G if n not in set(nodes)])
Subgraph views are sometimes NOT what you want. In most cases where you want to do more than simply look at the induced edges, it makes more sense to just create the subgraph as its own graph with code like:
# Create a subgraph SG based on a (possibly multigraph) G SG = G.__class__() SG.add_nodes_from((n, G.nodes[n]) for n in largest_wcc) if SG.is_multigraph(): SG.add_edges_from((n, nbr, key, d) for n, nbrs in G.adj.items() if n in largest_wcc for nbr, keydict in nbrs.items() if nbr in largest_wcc for key, d in keydict.items()) else: SG.add_edges_from((n, nbr, d) for n, nbrs in G.adj.items() if n in largest_wcc for nbr, d in nbrs.items() if nbr in largest_wcc) SG.graph.update(G.graph)
Examples
>>> G = nx.path_graph(4) # or DiGraph, MultiGraph, MultiDiGraph, etc >>> H = G.subgraph([0, 1, 2]) >>> list(H.edges) [(0, 1), (1, 2)]
-
class
wbia.algo.graph.nx_dynamic_graph.NiceGraph(incoming_graph_data=None, **attr)[source]¶ Bases:
networkx.classes.graph.Graph,wbia.algo.graph.nx_dynamic_graph.GraphHelperMixin
wbia.algo.graph.nx_edge_augmentation module¶
Algorithms for finding k-edge-augmentations
A k-edge-augmentation is a set of edges, that once added to a graph, ensures that the graph is k-edge-connected. Typically, the goal is to find the augmentation with minimum weight. In general, it is not gaurenteed that a k-edge-augmentation exists.
-
class
wbia.algo.graph.nx_edge_augmentation.MetaEdge(meta_uv, uv, w)¶ Bases:
tuple-
meta_uv¶ Alias for field number 0
-
uv¶ Alias for field number 1
-
w¶ Alias for field number 2
-
-
wbia.algo.graph.nx_edge_augmentation.bridge_augmentation(G, avail=None, weight=None)[source]¶ Finds the a set of edges that bridge connects G.
Adding these edges to G will make it 2-edge-connected. If no constraints are specified the returned set of edges is minimum an optimal, otherwise the solution is approximated.
Notes
If there are no constraints the solution can be computed in linear time using
unconstrained_bridge_augmentation(). Otherwise, the problem becomes NP-hard and is the solution is approximated byweighted_bridge_augmentation().
-
wbia.algo.graph.nx_edge_augmentation.collapse(G, grouped_nodes)[source]¶ Collapses each group of nodes into a single node.
This is similar to condensation, but works on undirected graphs.
Parameters: - G (NetworkX Graph) – A directed graph.
- grouped_nodes (list or generator) – Grouping of nodes to collapse. The grouping must be disjoint. If grouped_nodes are strongly_connected_components then this is equivalent to condensation.
Returns: C – The collapsed graph C of G with respect to the node grouping. The node labels are integers corresponding to the index of the component in the list of strongly connected components of G. C has a graph attribute named ‘mapping’ with a dictionary mapping the original nodes to the nodes in C to which they belong. Each node in C also has a node attribute ‘members’ with the set of original nodes in G that form the group that the node in C represents.
Return type: NetworkX Graph
Examples
>>> # Collapses a graph using disjoint groups, but not necesarilly connected >>> G = nx.Graph([(1, 0), (2, 3), (3, 1), (3, 4), (4, 5), (5, 6), (5, 7)]) >>> G.add_node('A') >>> grouped_nodes = [{0, 1, 2, 3}, {5, 6, 7}] >>> C = collapse(G, grouped_nodes) >>> members = nx.get_node_attributes(C, 'members') >>> sorted(members.keys()) [0, 1, 2, 3] >>> member_values = set(map(frozenset, members.values())) >>> assert {0, 1, 2, 3} in member_values >>> assert {4} in member_values >>> assert {5, 6, 7} in member_values >>> assert {'A'} in member_values
-
wbia.algo.graph.nx_edge_augmentation.complement_edges(G)[source]¶ Returns only the edges in the complement of G
Example
>>> G = nx.path_graph((1, 2, 3, 4)) >>> sorted(complement_edges(G)) [(1, 3), (1, 4), (2, 4)] >>> G = nx.path_graph((1, 2, 3, 4), nx.DiGraph()) >>> sorted(complement_edges(G)) [(1, 3), (1, 4), (2, 1), (2, 4), (3, 1), (3, 2), (4, 1), (4, 2), (4, 3)] >>> G = nx.complete_graph(1000) >>> sorted(complement_edges(G)) []
-
wbia.algo.graph.nx_edge_augmentation.greedy_k_edge_augmentation(G, k, avail=None, weight=None, seed=None)[source]¶ Greedy algorithm for finding a k-edge-augmentation
Notes
The algorithm is simple. Edges are incrementally added between parts of the graph that are not yet locally k-edge-connected. Then edges are from the augmenting set are pruned as long as local-edge-connectivity is not broken.
This algorithm is greedy and does not provide optimiality gaurentees. It exists only to provide
k_edge_augmentation()with the ability to generate a feasible solution for arbitrary k.Example
>>> G = nx.path_graph((1, 2, 3, 4, 5, 6, 7)) >>> sorted(greedy_k_edge_augmentation(G, k=2)) [(1, 7)] >>> sorted(greedy_k_edge_augmentation(G, k=1, avail=[])) [] >>> G = nx.path_graph((1, 2, 3, 4, 5, 6, 7)) >>> avail = {(u, v): 1 for (u, v) in complement_edges(G)} >>> # randomized pruning process can produce different solutions >>> sorted(greedy_k_edge_augmentation(G, k=4, avail=avail, seed=2)) [(1, 3), (1, 4), (1, 5), (1, 6), (1, 7), (2, 4), (2, 6), (3, 7), (5, 7)] >>> sorted(greedy_k_edge_augmentation(G, k=4, avail=avail, seed=3)) [(1, 3), (1, 5), (1, 6), (2, 4), (2, 6), (3, 7), (4, 7), (5, 7)]
-
wbia.algo.graph.nx_edge_augmentation.is_k_edge_connected(G, k)[source]¶ Tests to see if a graph is k-edge-connected
See also
Example
>>> G = nx.barbell_graph(10, 0) >>> is_k_edge_connected(G, k=1) True >>> is_k_edge_connected(G, k=2) False
-
wbia.algo.graph.nx_edge_augmentation.is_locally_k_edge_connected(G, s, t, k)[source]¶ Tests to see if an edge in a graph is locally k-edge-connected
See also
Example
>>> G = nx.barbell_graph(10, 0) >>> is_locally_k_edge_connected(G, 5, 15, k=1) True >>> is_locally_k_edge_connected(G, 5, 15, k=2) False >>> is_locally_k_edge_connected(G, 1, 5, k=2) True
-
wbia.algo.graph.nx_edge_augmentation.k_edge_augmentation(G, k, avail=None, weight=None, partial=False)[source]¶ Finds set of edges to k-edge-connect G.
This function uses the most efficient function available (depending on the value of k and if the problem is weighted or unweighted) to search for a minimum weight subset of available edges that k-edge-connects G. In general, finding a k-edge-augmentation is NP-hard, so solutions are not garuenteed to be minimal.
Parameters: - G (NetworkX graph) –
- k (Integer) – Desired edge connectivity
- avail (dict or a set 2 or 3 tuples) –
The available edges that can be used in the augmentation.
If unspecified, then all edges in the complement of G are available. Otherwise, each item is an available edge (with an optinal weight).
In the unweighted case, each item is an edge
(u, v).In the weighted case, each item is a 3-tuple
(u, v, d)or a dict with items(u, v): d. The third item,d, can be a dictionary or a real number. Ifdis a dictionaryd[weight]correspondings to the weight. - weight (string) – key to use to find weights if avail is a set of 3-tuples where the third item in each tuple is a dictionary.
- partial (Boolean) – If partial is True and no feasible k-edge-augmentation exists, then all available edges are returned.
Returns: aug_edges – the G would become k-edge-connected. If partial is False, an error is raised if this is not possible. Otherwise, all available edges are generated.
Return type: a generator of edges. If these edges are added to G, then
Raises: - NetworkXNotImplemented: – If the input graph is directed or a multigraph.
- ValueError: – If k is less than 1
Notes
When k=1 this returns an optimal solution.
When k=2 and avail is None, this returns an optimal solution. Otherwise when k=2, this returns a 2-approximation of the optimal solution.
- For k>3, this problem is NP-hard and this uses a randomized algorithm that
- produces a feasible solution, but provides no gaurentees on the solution weight.
Example
>>> # Unweighted cases >>> G = nx.path_graph((1, 2, 3, 4)) >>> G.add_node(5) >>> sorted(k_edge_augmentation(G, k=1)) [(1, 5)] >>> sorted(k_edge_augmentation(G, k=2)) [(1, 5), (5, 4)] >>> sorted(k_edge_augmentation(G, k=3)) [(1, 4), (1, 5), (2, 5), (3, 5), (4, 5)] >>> complement = list(k_edge_augmentation(G, k=5, partial=True)) >>> G.add_edges_from(complement) >>> nx.edge_connectivity(G) 4
Example
>>> # Weighted cases >>> G = nx.path_graph((1, 2, 3, 4)) >>> G.add_node(5) >>> # avail can be a tuple with a dict >>> avail = [(1, 5, {'weight': 11}), (2, 5, {'weight': 10})] >>> sorted(k_edge_augmentation(G, k=1, avail=avail, weight='weight')) [(2, 5)] >>> # or avail can be a 3-tuple with a real number >>> avail = [(1, 5, 11), (2, 5, 10), (4, 3, 1), (4, 5, 51)] >>> sorted(k_edge_augmentation(G, k=2, avail=avail)) [(1, 5), (2, 5), (4, 5)] >>> # or avail can be a dict >>> avail = {(1, 5): 11, (2, 5): 10, (4, 3): 1, (4, 5): 51} >>> sorted(k_edge_augmentation(G, k=2, avail=avail)) [(1, 5), (2, 5), (4, 5)] >>> # If augmentation is infeasible, then all edges in avail are returned >>> avail = {(1, 5): 11} >>> sorted(k_edge_augmentation(G, k=2, avail=avail, partial=True)) [(1, 5)]
-
wbia.algo.graph.nx_edge_augmentation.one_edge_augmentation(G, avail=None, weight=None, partial=False)[source]¶ Finds minimum weight set of edges to connect G.
Notes
Uses either
unconstrained_one_edge_augmentation()orweighted_one_edge_augmentation()depending on whetheravailis specified. Both algorithms are based on finding a minimum spanning tree. As such both algorithms find optimal solutions and run in linear time.
-
wbia.algo.graph.nx_edge_augmentation.partial_k_edge_augmentation(G, k, avail, weight=None)[source]¶ Finds augmentation that k-edge-connects as much of the graph as possible
When a k-edge-augmentation is not possible, we can still try to find a small set of edges that partially k-edge-connects as much of the graph as possible.
Notes
Construct H that augments G with all edges in avail. Find the k-edge-subgraphs of H. For each k-edge-subgraph, if the number of nodes is more than k, then find the k-edge-augmentation of that graph and add it to the solution. Then add all edges in avail between k-edge subgraphs to the solution.
>>> G = nx.path_graph((1, 2, 3, 4, 5, 6, 7)) >>> G.add_node(8) >>> avail = [(1, 3), (1, 4), (1, 5), (2, 4), (2, 5), (3, 5), (1, 8)] >>> sorted(partial_k_edge_augmentation(G, k=2, avail=avail)) [(1, 5), (1, 8)]
-
wbia.algo.graph.nx_edge_augmentation.unconstrained_bridge_augmentation(G)[source]¶ Finds an optimal 2-edge-augmentation of G using the fewest edges.
This is an implementation of the algorithm detailed in [1]_. The basic idea is to construct a meta-graph of bridge-ccs, connect leaf nodes of the trees to connect the entire graph, and finally connect the leafs of the tree in dfs-preorder to bridge connect the entire graph.
Notes
Input: a graph G. First find the bridge components of G and collapse each bridge-cc into a node of a metagraph graph C, which is gaurenteed to be a forest of trees.
C contains p “leafs” — nodes with exactly one incident edge. C contains q “isolated nodes” — nodes with no incident edges.
- Theorem: If p + q > 1, then at least
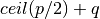 edges are
edges are - needed to bridge connect C. This algorithm achieves this min number.
The method first adds enough edges to make G into a tree and then pairs leafs in a simple fashion.
Let n be the number of trees in C. Let v(i) be an isolated vertex in the i-th tree if one exists, otherwise it is a pair of distinct leafs nodes in the i-th tree. Alternating edges from these sets (i.e. adding edges A1 = [(v(i)[0], v(i + 1)[1]), v(i + 1)[0], v(i + 2)[1])…]) connects C into a tree T. This tree has p’ = p + 2q - 2(n -1) leafs and no isolated vertices. A1 has n - 1 edges. The next step finds ceil(p’ / 2) edges to biconnect any tree with p’ leafs.
Convert T into an arborescence T’ by picking an arbitrary root node with degree >= 2 and directing all edges away from the root. Note the implementation implicitly constructs T’.
The leafs of T are the nodes with no existing edges in T’. Order the leafs of T’ by DFS prorder. Then break this list in half and add the zipped pairs to A2.
The set A = A1 + A2 is the minimum augmentation in the metagraph.
To convert this to edges in the original graph
References
[1] Eswaran, Kapali P., and R. Endre Tarjan. (1975) Augmentation problems. http://epubs.siam.org/doi/abs/10.1137/0205044 Example
>>> G = nx.path_graph((1, 2, 3, 4, 5, 6, 7)) >>> sorted(unconstrained_bridge_augmentation(G)) [(1, 7)] >>> G = nx.path_graph((1, 2, 3, 2, 4, 5, 6, 7)) >>> sorted(unconstrained_bridge_augmentation(G)) [(1, 3), (3, 7)] >>> G = nx.Graph([(0, 1), (0, 2), (1, 2)]) >>> G.add_node(4) >>> sorted(unconstrained_bridge_augmentation(G)) [(1, 4), (4, 0)]
- Theorem: If p + q > 1, then at least
-
wbia.algo.graph.nx_edge_augmentation.unconstrained_one_edge_augmentation(G)[source]¶ Finds the smallest set of edges to connect G.
This is a variant of the unweighted MST problem. If G is not empty, a feasible solution always exists.
Example
>>> G = nx.Graph([(1, 2), (2, 3), (4, 5)]) >>> G.add_nodes_from([6, 7, 8]) >>> sorted(unconstrained_one_edge_augmentation(G)) [(1, 4), (4, 6), (6, 7), (7, 8)]
-
wbia.algo.graph.nx_edge_augmentation.weighted_bridge_augmentation(G, avail, weight=None)[source]¶ Finds an approximate min-weight 2-edge-augmentation of G.
This is an implementation of the approximation algorithm detailed in [1]_. It chooses a set of edges from avail to add to G that renders it 2-edge-connected if such a subset exists. This is done by finding a minimum spanning arborescence of a specially constructed metagraph.
Parameters: - G (NetworkX graph) –
- avail (set of 2 or 3 tuples.) – candidate edges (with optional weights) to choose from
- weight (string) – key to use to find weights if avail is a set of 3-tuples where the third item in each tuple is a dictionary.
Returns: aug_edges (set)
Return type: subset of avail chosen to augment G
Notes
Finding a weighted 2-edge-augmentation is NP-hard. Any edge not in
availis considered to have a weight of infinity. The approximation factor is 2 ifGis connected and 3 if it is not. Runs in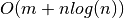 time
timeReferences
[1] Khuller, Samir, and Ramakrishna Thurimella. (1993) Approximation algorithms for graph augmentation. http://www.sciencedirect.com/science/article/pii/S0196677483710102 Example
>>> G = nx.path_graph((1, 2, 3, 4)) >>> # When the weights are equal, (1, 4) is the best >>> avail = [(1, 4, 1), (1, 3, 1), (2, 4, 1)] >>> sorted(weighted_bridge_augmentation(G, avail)) [(1, 4)] >>> # Giving (1, 4) a high weight makes the two edge solution the best. >>> avail = [(1, 4, 1000), (1, 3, 1), (2, 4, 1)] >>> sorted(weighted_bridge_augmentation(G, avail)) [(1, 3), (2, 4)] >>> #------ >>> G = nx.path_graph((1, 2, 3, 4)) >>> G.add_node(5) >>> avail = [(1, 5, 11), (2, 5, 10), (4, 3, 1), (4, 5, 1)] >>> sorted(weighted_bridge_augmentation(G, avail=avail)) [(1, 5), (4, 5)] >>> avail = [(1, 5, 11), (2, 5, 10), (4, 3, 1), (4, 5, 51)] >>> sorted(weighted_bridge_augmentation(G, avail=avail)) [(1, 5), (2, 5), (4, 5)]
-
wbia.algo.graph.nx_edge_augmentation.weighted_one_edge_augmentation(G, avail, weight=None, partial=False)[source]¶ Finds the minimum weight set of edges to connect G if one exists.
This is a variant of the weighted MST problem.
Example
>>> G = nx.Graph([(1, 2), (2, 3), (4, 5)]) >>> G.add_nodes_from([6, 7, 8]) >>> # any edge not in avail has an implicit weight of infinity >>> avail = [(1, 3), (1, 5), (4, 7), (4, 8), (6, 1), (8, 1), (8, 2)] >>> sorted(weighted_one_edge_augmentation(G, avail)) [(1, 5), (4, 7), (6, 1), (8, 1)] >>> # find another solution by giving large weights to edges in the >>> # previous solution (note some of the old edges must be used) >>> avail = [(1, 3), (1, 5, 99), (4, 7, 9), (6, 1, 99), (8, 1, 99), (8, 2)] >>> sorted(weighted_one_edge_augmentation(G, avail)) [(1, 5), (4, 7), (6, 1), (8, 2)]
wbia.algo.graph.nx_edge_kcomponents module¶
Algorithms for finding k-edge-connected components and subgraphs.
A k-edge-connected component (k-edge-cc) is a maximal set of nodes in G, such that all pairs of node have an edge-connectivity of at least k.
A k-edge-connected subgraph (k-edge-subgraph) is a maximal set of nodes in G, such that the subgraph of G defined by the nodes has an edge-connectivity at least k.
-
class
wbia.algo.graph.nx_edge_kcomponents.EdgeComponentAuxGraph[source]¶ Bases:
objectA simple algorithm to find all k-edge-connected components in a graph.
Constructing the AuxillaryGraph (which may take some time) allows for the k-edge-ccs to be found in linear time for arbitrary k.
Notes
This implementation is based on [1]_. The idea is to construct an auxillary graph from which the k-edge-ccs can be extracted in linear time. The auxillary graph is constructed in O(|V|F) operations, where F is the complexity of max flow. Querying the components takes an additional O(|V|) operations. This algorithm can be slow for large graphs, but it handles an arbitrary k and works for both directed and undirected inputs.
The undirected case for k=1 is exactly connected components. The undirected case for k=2 is exactly bridge connected components. The directed case for k=1 is exactly strongly connected components.
References
[1] Wang, Tianhao, et al. (2015) A simple algorithm for finding all k-edge-connected components. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0136264 Example
>>> from networkx.utils import pairwise >>> # Build an interesting graph with multiple levels of k-edge-ccs >>> paths = [ ... (1, 2, 3, 4, 1, 3, 4, 2), # a 3-edge-cc (a 4 clique) ... (5, 6, 7, 5), # a 2-edge-cc (a 3 clique) ... (1, 5), # combine first two ccs into a 1-edge-cc ... (0,), # add an additional disconnected 1-edge-cc ... ] >>> G = nx.Graph() >>> G.add_nodes_from(it.chain(*paths)) >>> G.add_edges_from(it.chain(*[pairwise(path) for path in paths])) >>> # Constructing the AuxGraph takes about O(n ** 4) >>> aux_graph = EdgeComponentAuxGraph.construct(G) >>> # Once constructed, querying takes O(n) >>> sorted(map(sorted, aux_graph.k_edge_components(k=1))) [[0], [1, 2, 3, 4, 5, 6, 7]] >>> sorted(map(sorted, aux_graph.k_edge_components(k=2))) [[0], [1, 2, 3, 4], [5, 6, 7]] >>> sorted(map(sorted, aux_graph.k_edge_components(k=3))) [[0], [1, 2, 3, 4], [5], [6], [7]] >>> sorted(map(sorted, aux_graph.k_edge_components(k=4))) [[0], [1], [2], [3], [4], [5], [6], [7]]
Example
>>> # The auxillary graph is primarilly used for k-edge-ccs but it >>> # can also speed up the queries of k-edge-subgraphs by refining the >>> # search space. >>> from networkx.utils import pairwise >>> paths = [ ... (1, 2, 4, 3, 1, 4), ... ] >>> G = nx.Graph() >>> G.add_nodes_from(it.chain(*paths)) >>> G.add_edges_from(it.chain(*[pairwise(path) for path in paths])) >>> aux_graph = EdgeComponentAuxGraph.construct(G) >>> sorted(map(sorted, aux_graph.k_edge_subgraphs(k=3))) [[1], [2], [3], [4]] >>> sorted(map(sorted, aux_graph.k_edge_components(k=3))) [[1, 4], [2], [3]]
-
classmethod
construct(G)[source]¶ Builds an auxillary graph encoding edge-connectivity between nodes.
Notes
Given G=(V, E), initialize an empty auxillary graph A. Choose an arbitrary source node s. Initialize a set N of available nodes (that can be used as the sink). The algorithm picks an arbitrary node t from N - {s}, and then computes the minimum st-cut (S, T) with value w. If G is directed the the minimum of the st-cut or the ts-cut is used instead. Then, the edge (s, t) is added to the auxillary graph with weight w. The algorithm is called recursively first using S as the available nodes and s as the source, and then using T and t. Recusion stops when the source is the only available node.
Parameters: G (NetworkX graph) –
-
k_edge_components(k)[source]¶ Queries the auxillary graph for k-edge-connected components.
Parameters: k (Integer) – Desired edge connectivity Returns: k_edge_components Return type: a generator of k-edge-ccs Notes
Given the auxillary graph, the k-edge-connected components can be determined in linear time by removing all edges with weights less than k from the auxillary graph. The resulting connected components are the k-edge-ccs in the original graph.
-
k_edge_subgraphs(k)[source]¶ Queries the auxillary graph for k-edge-connected subgraphs.
Parameters: k (Integer) – Desired edge connectivity Returns: k_edge_subgraphs Return type: a generator of k-edge-subgraphs Notes
Refines the k-edge-ccs into k-edge-subgraphs. The running time is more than O(|V|).
For single values of k it is faster to use nx.k_edge_subgraphs. But for multiple values of k, it can be faster to build AuxGraph and then use this method.
-
classmethod
-
wbia.algo.graph.nx_edge_kcomponents.bridge_components(G)[source]¶ Finds all bridge-connected components G.
Parameters: G (NetworkX undirected graph) – Returns: bridge_components Return type: a generator of 2-edge-connected components See also
k_edge_subgraphs()- this function is a special case for an undirected graph where k=2.
biconnected_components()- similar to this function, but is defined using 2-node-connectivity instead of 2-edge-connectivity.
Raises: NetworkXNotImplemented: – If the input graph is directed or a multigraph. Notes
Bridge-connected components are also known as 2-edge-connected components.
Example
>>> # The barbell graph with parameter zero has a single bridge >>> G = nx.barbell_graph(5, 0) >>> sorted(map(sorted, bridge_components(G))) [[0, 1, 2, 3, 4], [5, 6, 7, 8, 9]]
-
wbia.algo.graph.nx_edge_kcomponents.general_k_edge_subgraphs(G, k)[source]¶ General algorithm to find all maximal k-edge-connected subgraphs in G.
Returns: k_edge_subgraphs – Each k-edge-subgraph is a maximal set of nodes that defines a subgraph of G that is k-edge-connected. Return type: a generator of nx.Graphs that are k-edge-subgraphs Notes
Implementation of the basic algorithm from _[1]. The basic idea is to find a global minimum cut of the graph. If the cut value is at least k, then the graph is a k-edge-connected subgraph and can be added to the results. Otherwise, the cut is used to split the graph in two and the procedure is applied recursively. If the graph is just a single node, then it is also added to the results. At the end, each result is either gaurenteed to be a single node or a subgraph of G that is k-edge-connected.
This implementation contains optimizations for reducing the number of calls to max-flow, but there are other optimizations in _[1] that could be implemented.
References
[1] Zhou, Liu, et al. (2012) Finding maximal k-edge-connected subgraphs from a large graph. ACM International Conference on Extending Database Technology 2012 480-–491. https://openproceedings.org/2012/conf/edbt/ZhouLYLCL12.pdf Example
>>> from networkx.utils import pairwise >>> paths = [ ... (11, 12, 13, 14, 11, 13, 14, 12), # a 4-clique ... (21, 22, 23, 24, 21, 23, 24, 22), # another 4-clique ... # connect the cliques with high degree but low connectivity ... (50, 13), ... (12, 50, 22), ... (13, 102, 23), ... (14, 101, 24), ... ] >>> G = nx.Graph(it.chain(*[pairwise(path) for path in paths])) >>> sorted(map(len, k_edge_subgraphs(G, k=3))) [1, 1, 1, 4, 4]
-
wbia.algo.graph.nx_edge_kcomponents.k_edge_components(G, k)[source]¶ Generates nodes in each maximal k-edge-connected component in G.
Parameters: - G (NetworkX graph) –
- k (Integer) – Desired edge connectivity
Returns: k_edge_components – will have k-edge-connectivity in the graph G.
Return type: a generator of k-edge-ccs. Each set of returned nodes
See also
local_edge_connectivity()k_edge_subgraphs()- similar to this function, but the subgraph defined by the nodes must also have k-edge-connectivity.
k_components()- similar to this function, but uses node-connectivity instead of edge-connectivity
Raises: - NetworkXNotImplemented: – If the input graph is a multigraph.
- ValueError: – If k is less than 1
Notes
Attempts to use the most efficient implementation available based on k. If k=1, this is simply simply connected components for directed graphs and connected components for undirected graphs. If k=2 on an efficient bridge connected component algorithm from _[1] is run based on the chain decomposition. Otherwise, the algorithm from _[2] is used.
Example
>>> from networkx.utils import pairwise >>> paths = [ ... (1, 2, 4, 3, 1, 4), ... (5, 6, 7, 8, 5, 7, 8, 6), ... ] >>> G = nx.Graph() >>> G.add_nodes_from(it.chain(*paths)) >>> G.add_edges_from(it.chain(*[pairwise(path) for path in paths])) >>> # note this returns {1, 4} unlike k_edge_subgraphs >>> sorted(map(sorted, k_edge_components(G, k=3))) [[1, 4], [2], [3], [5, 6, 7, 8]]
References
[1] https://en.wikipedia.org/wiki/Bridge_(graph_theory) [2] Wang, Tianhao, et al. (2015) A simple algorithm for finding all k-edge-connected components. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0136264
-
wbia.algo.graph.nx_edge_kcomponents.k_edge_subgraphs(G, k)[source]¶ Generates nodes in each maximal k-edge-connected subgraph in G.
Parameters: - G (NetworkX graph) –
- k (Integer) – Desired edge connectivity
Returns: k_edge_subgraphs – Each k-edge-subgraph is a maximal set of nodes that defines a subgraph of G that is k-edge-connected.
Return type: a generator of k-edge-subgraphs
See also
edge_connectivity()k_edge_components()- similar to this function, but nodes only need to have k-edge-connctivity within the graph G and the subgraphs might not be k-edge-connected.
Raises: - NetworkXNotImplemented: – If the input graph is a multigraph.
- ValueError: – If k is less than 1
Notes
Attempts to use the most efficient implementation available based on k. If k=1, or k=2 and the graph is undirected, then this simply calls k_edge_components. Otherwise the algorithm from _[1] is used.
Example
>>> from networkx.utils import pairwise >>> paths = [ ... (1, 2, 4, 3, 1, 4), ... (5, 6, 7, 8, 5, 7, 8, 6), ... ] >>> G = nx.Graph() >>> G.add_nodes_from(it.chain(*paths)) >>> G.add_edges_from(it.chain(*[pairwise(path) for path in paths])) >>> # note this does not return {1, 4} unlike k_edge_components >>> sorted(map(sorted, k_edge_subgraphs(G, k=3))) [[1], [2], [3], [4], [5, 6, 7, 8]]
References
[1] Zhou, Liu, et al. (2012) Finding maximal k-edge-connected subgraphs from a large graph. ACM International Conference on Extending Database Technology 2012 480-–491. https://openproceedings.org/2012/conf/edbt/ZhouLYLCL12.pdf
wbia.algo.graph.nx_utils module¶
TODO: the k-components will soon be implemented in networkx 2.0 use those instead
-
wbia.algo.graph.nx_utils.demodata_tarjan_bridge()[source]¶ - CommandLine:
- python -m wbia.algo.graph.nx_utils demodata_tarjan_bridge –show
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_utils import * # NOQA >>> G = demodata_tarjan_bridge() >>> ut.quit_if_noshow() >>> import wbia.plottool as pt >>> pt.show_nx(G) >>> ut.show_if_requested()
-
wbia.algo.graph.nx_utils.diag_product(s1, s2)[source]¶ Does product, but iterates over the diagonal first
-
wbia.algo.graph.nx_utils.edges_between(graph, nodes1, nodes2=None, assume_disjoint=False, assume_dense=True)[source]¶ Get edges between two components or within a single component
Parameters: - CommandLine:
- python -m wbia.algo.graph.nx_utils –test-edges_between
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_utils import * # NOQA >>> import utool as ut >>> edges = [ >>> (1, 2), (2, 3), (3, 4), (4, 1), (4, 3), # cc 1234 >>> (1, 5), (7, 2), (5, 1), # cc 567 / 5678 >>> (7, 5), (5, 6), (8, 7), >>> ] >>> digraph = nx.DiGraph(edges) >>> graph = nx.Graph(edges) >>> nodes1 = [1, 2, 3, 4] >>> nodes2 = [5, 6, 7] >>> n2 = sorted(edges_between(graph, nodes1, nodes2)) >>> n4 = sorted(edges_between(graph, nodes1)) >>> n5 = sorted(edges_between(graph, nodes1, nodes1)) >>> n1 = sorted(edges_between(digraph, nodes1, nodes2)) >>> n3 = sorted(edges_between(digraph, nodes1)) >>> print('n2 == %r' % (n2,)) >>> print('n4 == %r' % (n4,)) >>> print('n5 == %r' % (n5,)) >>> print('n1 == %r' % (n1,)) >>> print('n3 == %r' % (n3,)) >>> assert n2 == ([(1, 5), (2, 7)]), '2' >>> assert n4 == ([(1, 2), (1, 4), (2, 3), (3, 4)]), '4' >>> assert n5 == ([(1, 2), (1, 4), (2, 3), (3, 4)]), '5' >>> assert n1 == ([(1, 5), (5, 1), (7, 2)]), '1' >>> assert n3 == ([(1, 2), (2, 3), (3, 4), (4, 1), (4, 3)]), '3' >>> n6 = sorted(edges_between(digraph, nodes1 + [6], nodes2 + [1, 2], assume_dense=False)) >>> print('n6 = %r' % (n6,)) >>> n6 = sorted(edges_between(digraph, nodes1 + [6], nodes2 + [1, 2], assume_dense=True)) >>> print('n6 = %r' % (n6,)) >>> assert n6 == ([(1, 2), (1, 5), (2, 3), (4, 1), (5, 1), (5, 6), (7, 2)]), '6'
-
wbia.algo.graph.nx_utils.edges_cross(graph, nodes1, nodes2)[source]¶ Finds edges between two sets of disjoint nodes. Running time is O(len(nodes1) * len(nodes2))
Parameters:
-
wbia.algo.graph.nx_utils.edges_inside(graph, nodes)[source]¶ Finds edges within a set of nodes Running time is O(len(nodes) ** 2)
Parameters: - graph (nx.Graph) – an undirected graph
- nodes1 (set) – a set of nodes
-
wbia.algo.graph.nx_utils.edges_outgoing(graph, nodes)[source]¶ Finds edges leaving a set of nodes. Average running time is O(len(nodes) * ave_degree(nodes)) Worst case running time is O(G.number_of_edges()).
Parameters: - graph (nx.Graph) – a graph
- nodes (set) – set of nodes
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.nx_utils import * # NOQA >>> import utool as ut >>> G = demodata_bridge() >>> nodes = {1, 2, 3, 4} >>> outgoing = edges_outgoing(G, nodes) >>> assert outgoing == {(3, 5), (4, 8)}
-
wbia.algo.graph.nx_utils.random_k_edge_connected_graph(size, k, p=0.1, rng=None)[source]¶ Super hacky way of getting a random k-connected graph
Example
>>> # ENABLE_DOCTEST >>> import wbia.plottool as pt >>> from wbia.algo.graph.nx_utils import * # NOQA >>> size, k, p = 25, 3, .1 >>> rng = ut.ensure_rng(0) >>> gs = [] >>> for x in range(4): >>> G = random_k_edge_connected_graph(size, k, p, rng) >>> gs.append(G) >>> ut.quit_if_noshow() >>> pnum_ = pt.make_pnum_nextgen(nRows=2, nSubplots=len(gs)) >>> fnum = 1 >>> for g in gs: >>> pt.show_nx(g, fnum=fnum, pnum=pnum_())
wbia.algo.graph.refresh module¶
-
class
wbia.algo.graph.refresh.RefreshCriteria(window=20, patience=72, thresh=0.1, method='binomial')[source]¶ Bases:
objectDetermine when to re-query for candidate edges.
Models an upper bound on the probability that any of the next patience reviews will be label-changing (meaningful). Once this probability is below a threshold the criterion triggers. The model is either binomial or poisson. They both work about the same. The binomial is a slightly better model.
Does this by maintaining an estimate of the probability any particular review will be label-chaging using an exponentially weighted moving average. This is the rate parameter / individual event probability.
-
ave(method='exp')[source]¶ >>> # ENABLE_DOCTEST >>> from wbia.algo.graph.refresh import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=40, size=4, size_std=2, ignore_pair=True) >>> edges = list(infr.dummy_verif.find_candidate_edges(K=100)) >>> scores = np.array(infr.dummy_verif.predict_edges(edges)) >>> #sortx = ut.shuffle(np.arange(len(edges)), rng=321) >>> sortx = scores.argsort()[::-1] >>> edges = ut.take(edges, sortx) >>> scores = scores[sortx] >>> ys = infr.match_state_df(edges)[POSTV].values >>> y_remainsum = ys[::-1].cumsum()[::-1] >>> refresh = RefreshCriteria(window=250) >>> ma1 = [] >>> ma2 = [] >>> reals = [] >>> xdata = [] >>> for count, (edge, y) in enumerate(zip(edges, ys)): >>> refresh.add(y, user_id='user:oracle') >>> ma1.append(refresh._ewma) >>> ma2.append(refresh.pos_frac) >>> n_real = y_remainsum[count] / (len(edges) - count) >>> reals.append(n_real) >>> xdata.append(count + 1) >>> ut.quit_if_noshow() >>> import wbia.plottool as pt >>> pt.qtensure() >>> pt.multi_plot(xdata, [ma1, ma2, reals], marker='', >>> label_list=['exp', 'win', 'real'], xlabel='review num', >>> ylabel='mu')
-
pos_frac¶
-
pred_num_positives(n_remain_edges)[source]¶ Uses poisson process to estimate remaining positive reviews.
Multipling mu * n_remain_edges gives a probabilistic upper bound on the number of errors remaning. This only provides a real estimate if reviewing in a random order
Example
>>> # ENABLE_DOCTEST >>> from wbia.algo.graph.refresh import * # NOQA >>> from wbia.algo.graph import demo >>> infr = demo.demodata_infr(num_pccs=50, size=4, size_std=2) >>> edges = list(infr.dummy_verif.find_candidate_edges(K=100)) >>> #edges = ut.shuffle(sorted(edges), rng=321) >>> scores = np.array(infr.dummy_verif.predict_edges(edges)) >>> sortx = scores.argsort()[::-1] >>> edges = ut.take(edges, sortx) >>> scores = scores[sortx] >>> ys = infr.match_state_df(edges)[POSTV].values >>> y_remainsum = ys[::-1].cumsum()[::-1] >>> refresh = RefreshCriteria(window=250) >>> n_pred_list = [] >>> n_real_list = [] >>> xdata = [] >>> for count, (edge, y) in enumerate(zip(edges, ys)): >>> refresh.add(y, user_id='user:oracle') >>> n_remain_edges = len(edges) - count >>> n_pred = refresh.pred_num_positives(n_remain_edges) >>> n_real = y_remainsum[count] >>> if count == 2000: >>> break >>> n_real_list.append(n_real) >>> n_pred_list.append(n_pred) >>> xdata.append(count + 1) >>> ut.quit_if_noshow() >>> import wbia.plottool as pt >>> pt.qtensure() >>> n_pred_list = n_pred_list[10:] >>> n_real_list = n_real_list[10:] >>> xdata = xdata[10:] >>> pt.multi_plot(xdata, [n_pred_list, n_real_list], marker='', >>> label_list=['pred', 'real'], xlabel='review num', >>> ylabel='pred remaining merges') >>> stop_point = xdata[np.where(y_remainsum[10:] == 0)[0][0]] >>> pt.gca().plot([stop_point, stop_point], [0, int(max(n_pred_list))], 'g-')
-
wbia.algo.graph.state module¶
Module contents¶
-
wbia.algo.graph.IMPORT_TUPLES= []¶ cd /home/joncrall/code/wbia/wbia/algo/graph makeinit.py –modname=wbia.algo.graph
Type: Regen Command
-
wbia.algo.graph.reassign_submodule_attributes(verbose=1)[source]¶ Updates attributes in the __init__ modules with updated attributes in the submodules.
-
wbia.algo.graph.rrrr(verbose=1)¶ Reloads wbia.algo.graph and submodules